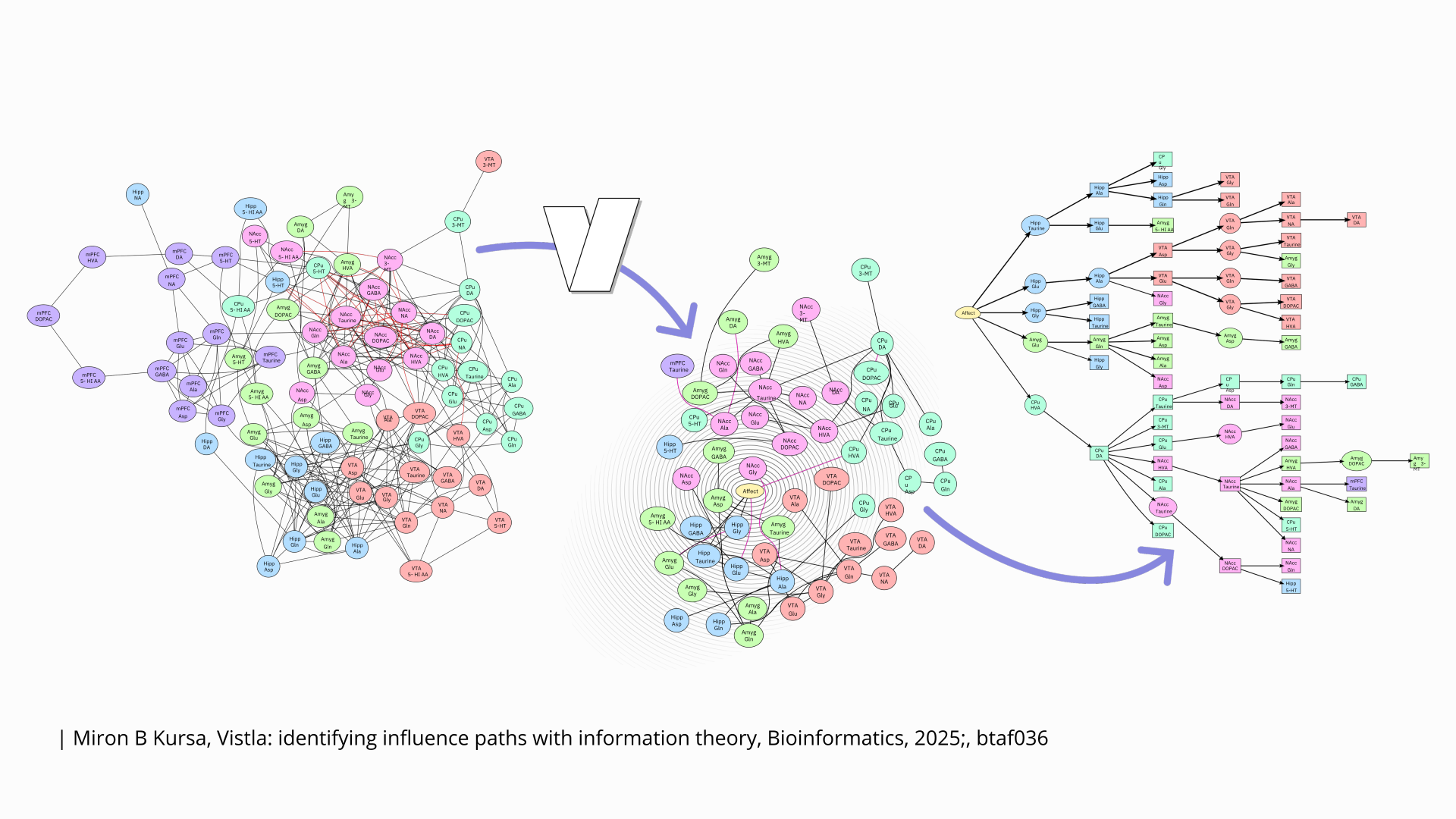
How to untangle a “haystack”?
An article by Dr. Miron Kursa from ICM UW titled “Vistla: identifying influence paths with information theory” has been published in Bioinformatics. It describes the Vistla algorithm, which is used for analyzing data from observations of complex systems, particularly those found in living organisms, studied through high-throughput molecular methods.
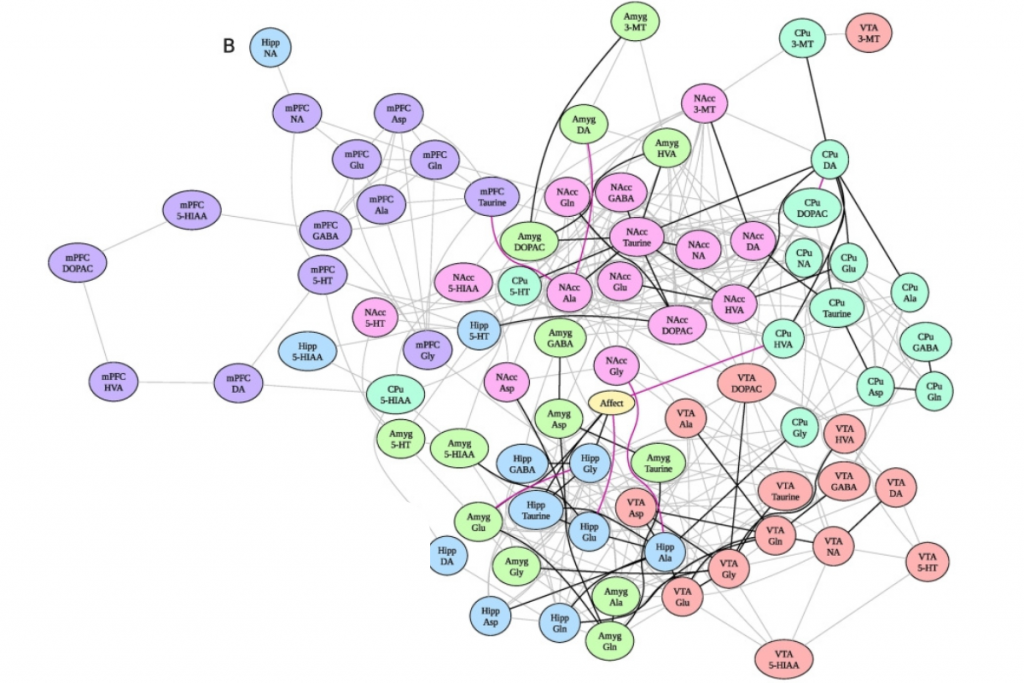
A complex system consists of components that interact with each other in a multitude of intricate and non-trivial ways. The network of these interactions, characterized by a high density of connections and, at first glance, a lack of clear structure, is commonly called a “haystack.” This complexity poses a significant challenge when attempting to decipher the specific mechanisms of the system’s behavior or when trying to steer its behavior toward a desired direction.
Vistla allows for the analysis of data from experiments in which the system was subjected to external intervention, such as the administration of a substance. The method tracks how the information about this event spreads through the system, passing between various components—resulting in a much easier-to-interpret graph with a tree-like topology.
The open-source implementation of the method for the R environment is available in the CRAN repository: https://doi.org/10.32614/CRAN.package.vistla